
Academic staff
Frédéric PECORARI
Permanent researcher (CNRS), group leaderCoordonnées
Institut de Recherche en Santé de l'Université de Nantes (IRS-UN) INSERM U1232 - CNRS ERL 6001 - CRCINA Équipe "Recherche en oncologie nucléaire" 8 quai Moncousu BP 70721 44007 Nantes cedex 1 Website: http://www.crcna.univ-nantes.fr .
- Tél
- 0228080268 (n° interne : 320268)
- Frederic.Pecorari@univ-nantes.fr
- Site internet
- http://intercept.cnrs.fr
Thèmes de recherche
Version Française
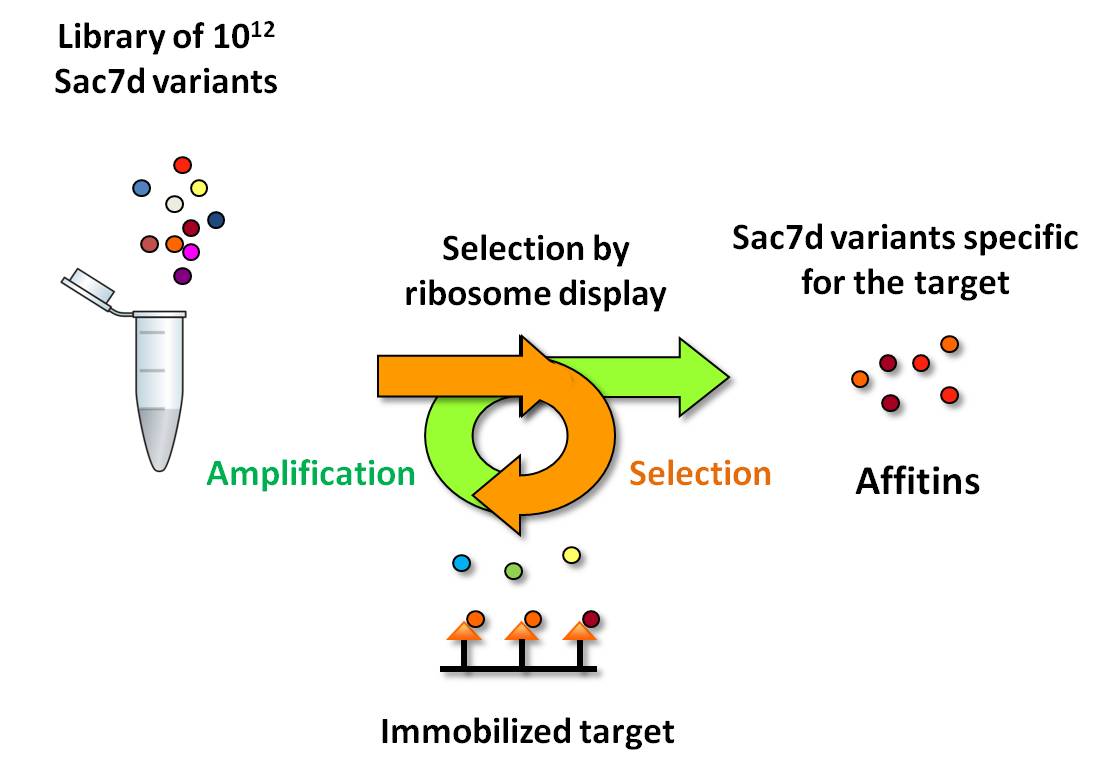
We are developing the use of polypeptides belonging to the archaeal Sul7d family, such as Sac7d, Sso7d and more recently Aho7c, as a non-antibody scaffold. Sul7d proteins combine a number of favorable properties from which artificial affinity proteins (Affitins) can be derived. Affitins are obtained by coupling the use of combinatorial libraries corresponding to the random mutation of amino acids at the surface of a Sul7d protein to in vitro selection against the target. Affitins show affinity and specificity that compare well with those of antibodies. They are thermally (up to 90°C) and chemically stable (pH, denaturants), well expressed in E. coli (up to 200 mg/L). With their SH3-like fold, they are structurally less complex than classical antibodies and 20 times smaller, a size compatible (7 kDa) with full chemical synthesis. We have demonstrated their use as reagent for intra-cellular inhibition, immunolocalization, protein chip array, biosensors, affinity chromatography and magnetic fishing applications. We also described at structural level how Affitins can inhibit enzymatic activities. We are now exploring their use for biomedical applications where fine tuning of the affinity reagent properties is needed.
We are developing the use of polypeptides belonging to the archaeal Sul7d family, such as Sac7d, Sso7d and more recently Aho7c, as a non-antibody scaffold. Sul7d proteins combine a number of favorable properties from which artificial affinity proteins (Affitins) can be derived. Affitins are obtained by coupling the use of combinatorial libraries corresponding to the random mutation of amino acids at the surface of a Sul7d protein to in vitro selection against the target. Affitins show affinity and specificity that compare well with those of antibodies. They are thermally (up to 90°C) and chemically stable (pH, denaturants), well expressed in E. coli (up to 200 mg/L). With their SH3-like fold, they are structurally less complex than classical antibodies and 20 times smaller, a size compatible (7 kDa) with full chemical synthesis. We have demonstrated their use as reagent for intra-cellular inhibition, immunolocalization, protein chip array, biosensors, affinity chromatography and magnetic fishing applications. We also described at structural level how Affitins can inhibit enzymatic activities. We are now exploring their use for biomedical applications where fine tuning of the affinity reagent properties is needed.
Informations complémentaires
Process for the generation of Affitins:
Patent application: PCT/IB2007/004388
Inventors : Frédéric Pecorari and Pedro Alzari (Institut Pasteur / CNRS)
Scientific publications related to Affitins:
A novel, smaller scaffold for Affitins: Showcase with binders specific for EpCAM
Kalichuk V, Renodon-Cornière A, Béhar G, Carrión F, Obal G, Maillasson M, Mouratou B, Préat V, Pecorari F.
Biotechnol Bioeng. 2017 Oct 4. doi: 10.1002/bit.26463
The archaeal "7kDa DNA-binding" proteins: extended characterization of an old gifted family
Kalichuk V, Béhar G, Renodon-Cornière A, Danovski G, Obal G, Barbet J, Mouratou B, Pecorari F.
Sci Rep. 2016 Nov 17;6:37274. doi: 10.1038/srep37274
Affitins for protein purification by affinity magnetic fishing
Fernandes CS, Dos Santos R, Ottengy S, Viecinski AC, Béhar G, Mouratou B, Pecorari F, Roque AC.
J. Chromatogr. A. 2016 Jun 7;S0021-9673(16)30779-8
Affitins as robust tailored reagents for affinity chromatography purification of antibodies and non-immunoglobulin proteins
Béhar G, Renodon-Cornière A, Mouratou B, Pecorari, F.
J. Chromatogr. A. 2016 Apr 8;1441, 44-51
Artificial Affinity Proteins as Ligands of Immunoglobulins
Mouratou B, Béhar G, Pecorari F.
Biomolecules. 2015 Jan 30;5(1):60-75
Switching an anti-IgG binding site between archaeal extremophilic proteins results in Affitins with enhanced pH stability
Béhar G, Pacheco S, Maillasson M, Mouratou B, Pecorari F.
J. Biotechnol, 2014, Dec;192, Part A(0):123-9.
Affinity transfer to the archaeal extremophilic Sac7d protein by insertion of a CDR
Pacheco S, Béhar G, Maillasson M, Mouratou B, Pecorari F.
Protein Eng Des Sel. 2014 Oct;27(10):431-8
Potent and specific inhibition of glycosidases by small artificial binding proteins (Affitins)
Correa A, Pacheco S, Mechaly AE, Obal G, Béhar G, Mouratou B, Oppezzo P, Alzari PM, Pecorari F.
PLoS One. 2014, 9(5):e97438
Tolerance of the archaeal Sac7d scaffold protein to alternative library designs: characterization of anti-immunoglobulin G Affitins
Behar G., Bellinzoni M., Maillasson M., Paillard-Laurance L., Alzari P.M., He X., Mouratou B. and Pecorari F.
Protein Eng Des Sel. 2013 Apr;26(4):267-75
Engineering of a phosphorylatable tag for specific protein binding on zirconium phosphonatebased microarrays
Cinier M, Petit M, Pecorari F, Talham DR, Bujoli B, Tellier C
J Biol Inorg Chem. 2012, 17(3):399-407
Reagentless fluorescent biosensors from artificial families of antigen binding proteins
Miranda FF, Brient-Litzler E, Zidane N, Pecorari F, Bedouelle H
Biosens Bioelectron. 2011, 26(10):4184-90
Bisphosphonate adaptors for specific protein binding on zirconium phosphonate-based microarrays
Cinier M, Petit M, Williams MN, Fabre RM, Pecorari F, Talham DR, Bujoli B, Tellier C
Bioconjugate Chemistry. 2009, 20(12): 2270-7
Type II secretion system secretin PulD localizes in clusters in the Escherichia coli outer membrane
Buddelmeijer N, Krehenbrink M, Pecorari F, Pugsley AP
J Bacteriol. 2009, 191(1):161-8
Artificial binding proteins (Affitins) as probes for conformational changes in secretin PulD
Krehenbrink M, Chami M, Guilvout I, Alzari PM, Pecorari F, Pugsley AP
J Mol Biol. 2008, 383(5):1058-68
Remodeling a DNA-binding protein as a specific in vivo inhibitor of bacterial secretin PulD
Mouratou B, Schaeffer F, Guilvout I, Tello-Manigne D, Pugsley AP, Alzari PM, Pecorari F
Proc Natl Acad Sci U S A. 2007, 104(46):17983-8
Patent application: PCT/IB2007/004388
Inventors : Frédéric Pecorari and Pedro Alzari (Institut Pasteur / CNRS)

Scientific publications related to Affitins:
A novel, smaller scaffold for Affitins: Showcase with binders specific for EpCAM
Kalichuk V, Renodon-Cornière A, Béhar G, Carrión F, Obal G, Maillasson M, Mouratou B, Préat V, Pecorari F.
Biotechnol Bioeng. 2017 Oct 4. doi: 10.1002/bit.26463
The archaeal "7kDa DNA-binding" proteins: extended characterization of an old gifted family
Kalichuk V, Béhar G, Renodon-Cornière A, Danovski G, Obal G, Barbet J, Mouratou B, Pecorari F.
Sci Rep. 2016 Nov 17;6:37274. doi: 10.1038/srep37274
Affitins for protein purification by affinity magnetic fishing
Fernandes CS, Dos Santos R, Ottengy S, Viecinski AC, Béhar G, Mouratou B, Pecorari F, Roque AC.
J. Chromatogr. A. 2016 Jun 7;S0021-9673(16)30779-8
Affitins as robust tailored reagents for affinity chromatography purification of antibodies and non-immunoglobulin proteins
Béhar G, Renodon-Cornière A, Mouratou B, Pecorari, F.
J. Chromatogr. A. 2016 Apr 8;1441, 44-51
Artificial Affinity Proteins as Ligands of Immunoglobulins
Mouratou B, Béhar G, Pecorari F.
Biomolecules. 2015 Jan 30;5(1):60-75
Switching an anti-IgG binding site between archaeal extremophilic proteins results in Affitins with enhanced pH stability
Béhar G, Pacheco S, Maillasson M, Mouratou B, Pecorari F.
J. Biotechnol, 2014, Dec;192, Part A(0):123-9.
Affinity transfer to the archaeal extremophilic Sac7d protein by insertion of a CDR
Pacheco S, Béhar G, Maillasson M, Mouratou B, Pecorari F.
Protein Eng Des Sel. 2014 Oct;27(10):431-8
Potent and specific inhibition of glycosidases by small artificial binding proteins (Affitins)
Correa A, Pacheco S, Mechaly AE, Obal G, Béhar G, Mouratou B, Oppezzo P, Alzari PM, Pecorari F.
PLoS One. 2014, 9(5):e97438
Tolerance of the archaeal Sac7d scaffold protein to alternative library designs: characterization of anti-immunoglobulin G Affitins
Behar G., Bellinzoni M., Maillasson M., Paillard-Laurance L., Alzari P.M., He X., Mouratou B. and Pecorari F.
Protein Eng Des Sel. 2013 Apr;26(4):267-75
Engineering of a phosphorylatable tag for specific protein binding on zirconium phosphonatebased microarrays
Cinier M, Petit M, Pecorari F, Talham DR, Bujoli B, Tellier C
J Biol Inorg Chem. 2012, 17(3):399-407
Reagentless fluorescent biosensors from artificial families of antigen binding proteins
Miranda FF, Brient-Litzler E, Zidane N, Pecorari F, Bedouelle H
Biosens Bioelectron. 2011, 26(10):4184-90
Bisphosphonate adaptors for specific protein binding on zirconium phosphonate-based microarrays
Cinier M, Petit M, Williams MN, Fabre RM, Pecorari F, Talham DR, Bujoli B, Tellier C
Bioconjugate Chemistry. 2009, 20(12): 2270-7
Type II secretion system secretin PulD localizes in clusters in the Escherichia coli outer membrane
Buddelmeijer N, Krehenbrink M, Pecorari F, Pugsley AP
J Bacteriol. 2009, 191(1):161-8
Artificial binding proteins (Affitins) as probes for conformational changes in secretin PulD
Krehenbrink M, Chami M, Guilvout I, Alzari PM, Pecorari F, Pugsley AP
J Mol Biol. 2008, 383(5):1058-68
Remodeling a DNA-binding protein as a specific in vivo inhibitor of bacterial secretin PulD
Mouratou B, Schaeffer F, Guilvout I, Tello-Manigne D, Pugsley AP, Alzari PM, Pecorari F
Proc Natl Acad Sci U S A. 2007, 104(46):17983-8
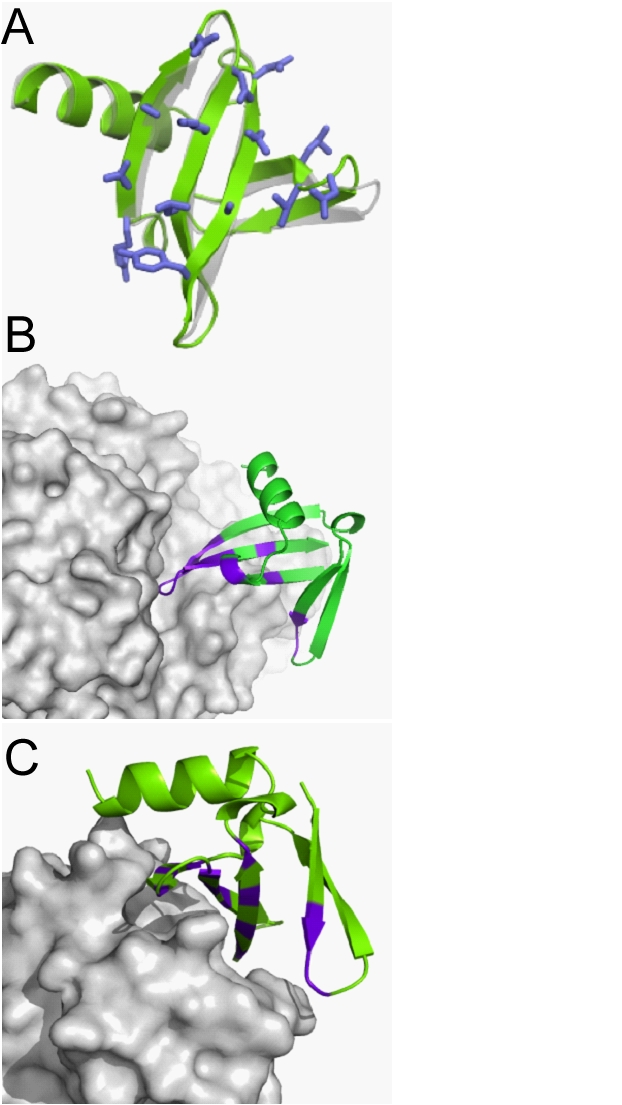
Affitins
A) First experimental structure of an Affitin (green, pdb entry 2XIW) superimposed with the structure of one of the molecular basis of artificial affinity proteins , here Sac7d (gray, pdb entry 1WTP). B) et C) Structures of two Affitins (green) in complex with enzymes (gray). These Affitins showed potent and specific inhibition of two glycosidases by covering or entering into catalytic sites. PDB codes: 4J0, 4CJ2. This alternative to antibodies combines favorable properties for biomedical applications we explore in the "Nuclear Oncology Research" team.
Mis à jour le 18 février 2025.
